| ♣ |
What is HUP-ML? |
♣ |
HUP-ML (Human Proteome Markup Language) is a XML based and proteomics-oriented markup language for exchanging proteome data between researchers to accelerate their collaboration. HUP-ML contains not only the details of methodology and experimental conditions, but also the results of proteome analysis. This feature helps with the precision check and comparison of experimental data which are obtained from different labs.
|
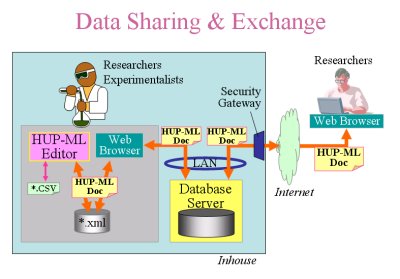 |
XML is the data format that is human readable and highly flexible because of tree structure. XML is playing an increasingly important role in the exchange of a wide variety of data on the Web. Additionally, the XML handling tools based on a standardized specification by W3C is available for any software platforms and is becoming very powerful to develop databases or software. Each form of XML data format is usually defined by document type definition (DTD) or Schema that tells what data will appear in. Please check the download site to download a DTD of HUP-ML. Comparing with other proteomics oriented XML formats, HUP-ML can describe in more detail at the point of experimental conditions and the results of protein identification. Therefore, describing proteomics data as following HUP-ML suggestion makes the proteome data linkable easily to other databases of life science such as genomics databases. HUP-ML does not contain clinical information of sources because of ethical issues in terms of privacy protection of volunteers.
|
|
| ♣ |
The details of HUP-ML |
♣ |
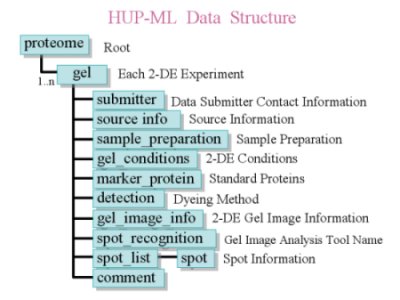
| The preparation parameter in the gel element consists of submitters, source information, sample preparation, gel conditions, marker proteins, detection method of gel spot, gel image and spot recognition. Additionally, the results of 2-dimensional electrophoresis experiments (2-DE) analysis can be stored in spot list elements under the gel element with capability of multiple storage of the results. Each spot element can explain spot location, information of protein identification including raw data of mass spectrum (MS) and identify protein data such as protein name, gene data and list of references. Currently, HUP-ML incorporates 2-DE analysis. The liquid chromatography profiles will be adopted next. |
|
|
| ♣ |
HUP-ML Editor Version 0.80 |
♣ |
HUP-ML Editor can manage HUP-ML without any XML knowledge.
HUP-ML Editor Version 0.80 features:
- Graphical user interface to create and edit HUP-ML formatted data.
- Template functions to avoid re-entry of the same information.
- Clickable 2-DE gel image viewer and print function.
- One step viewer of public data base through the accession number acquired by protein identification.
- Protein data importing function to merge related data from public data base.
- MS raw data importing and charting function.
- Automated data conversion from older version of HUP-ML to current one.
Please check the download site to download HUP-ML editor as well.
|
|
| ♣ |
References |
♣ |
- K. Kamijo et al. "A Proposition of XML Format for Proteomics Database", Proc. Of 18th International CODATA Conference, p.50, 2002.
- H. Mizuguchi et al. "An XML Format for Proteomics Database to Accelerate Collaboration among Bioresearchers", Journal of Japan Society of Information and knowledge, Vol.12 No.4 22-31, 2003.
- K. Kamijo et al. proposed HUP-ML in AOHUPO XML Standards Initiative at AOHUPO XML Workshop, 19 Dec. 2002.
- A. Kenmochi et al. proposes HUP-ML at the HUPO 2nd Annual & IUBMB XIX Joint World Congress, 8-11 Oct. 2003.
|
|

